The SAIN (Small Animal Imaging Network), which brings together scientists involved in small animal imaging at the national level, organizes Advanced Seminars in Small Animal In Vivo Imaging (SAIPAs).
SAIPAs consist of a series of webinars on three topics:
S1: Development of advanced MRI image acquisition sequences. Leaders: Emeline Ribot & Aurélien Trottier (RMSB, Bordeaux), Denis Grenier (Creatis, Lyon) + a Bruker representative.
S2: Image preprocessing and visualization. Leaders: Benjamin Lemasson (GIN, Grenoble), Sébastien Mériaux (NeuroSpin, Saclay), Sorina Pop (Créatis Lyon).
S3: Machine learning-based analysis methods. Leaders: Benjamin Lemasson (GIN, Grenoble), Vincent Noblet (Icube, Strasbourg), Sorina Pop (Créatis Lyon).
The target audience is students and researchers involved in projects based on small animal imaging and interested in advanced techniques for acquiring, processing, and analyzing these images.
The SAIPAs have multiple objectives: to promote and disseminate the expertise of SAIN group members, to train researchers in advanced techniques in the field of small animal in vivo imaging, and to raise awareness among participants about sharing data and tools in this field (compliance with the 3Rs policy, open data, traceability of treatments). SAIPAs will benefit from the FLI-IAM infrastructure for data management and pipeline execution.
This first webinar on the topic of S3: Machine learning-based analysis methods isscheduled for November 10, 2022 (details below).
The aim of this webinar is to demonstrate, through interactive practical exercises, the use of different convolutional neural networks applied to preclinical MRI imaging data. This webinar will focus on two use cases: i) artifact correction and ii) image classification.
Description: Presentation and introduction to MONAI [1], a Python-based development environment for artificial intelligence algorithms, through two practical cases using preclinical MRI data: i) artifact correction and ii) image classification.
Artifact correction:
Motion artifacts are a recurring problem in MRI and can manifest themselves in various ways on reconstructed images. For this practical assignment, we will work on artifacts visible in brain imaging that are most often related to the respiratory movements of animals whose heads are not properly secured in the cradle:
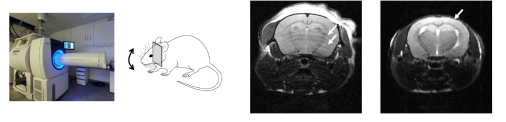
We will see how to simulate these motion artifacts in order to create a large dataset for training a convolutional neural network based on learning the residuals between the artifacted image and the artifact-free image [2]. The objective of this practical exercise is to show how we can use convolutional neural networks to generate corrected images from images containing motion artifacts.
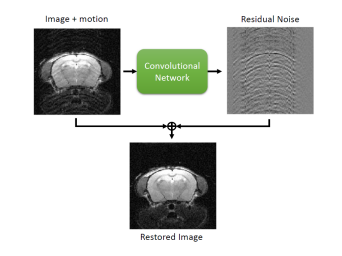
Image classification:
Image classification is one of the most common tasks performed by neural networks. In this second practical case, we will learn how to use the tools needed to train a convolutional neural network capable of automatically detecting the presence of a brain tumor or stroke from multiparametric MRI images in rats. This practical exercise will be an opportunity to review the different hyperparameters of a convolutional neural network as well as the data augmentation essential for their training. Finally, we will present a method called “Grad-CAM” which allows us to visualize the parts of the image that are most important for its classification [3].
Hours: Half day (November 10, 2022; 1:30 p.m. to 5:30 p.m.)
Prerequisites: Since we will be using the Google environment to i) share image datasets and ii) run notebooks on Google Colab, you will need a Google account and Google Chrome installed on your computer. If you do not have a Google account (or do not wish to share your personal email address), you can easily create an account dedicated to this seminar. Knowledge of Python is a plus, but not essential. Those who wish to familiarize themselves with the MONAI environment before this webinar can do so via these tutorials available on MONAI’s Github [4-5].
Registration : https://survey.creatis.insa-lyon.fr/index.php/356311?lang=fr
Presenters/organizers:
Benjamin Lemasson (CR, Inserm, GIN, Grenoble),
Mikaël Naveau (IR, Cyceron, Caen)
Vincent Noblet (IR, CNRS, ICube, Strasbourg)
Sorina Pop (IR, CNRS, Creatis, Lyon)
Franck Valentini (PhD student, University of Strasbourg, ICube, Strasbourg),
References :
[1] https://monai.io/
[2] Beyong a Gaussian Denoiser : Residual Learning of Deep CNN for Image Denoising, Zhang et al.,
2016
[3] Grad-CAM: Visual Explanations from Deep Networks via: Gradient-based Localization. R. R.
Selvaraju et al. IEEE; 2017
[4] https://github.com/Project-MONAI/tutorials/blob/main/3d_classification/densenet_training_a
rray.ipynb
[5] https://github.com/Project-MONAI/tutorials/blob/main/3d_segmentation/swin_unetr_brats21




